Laboratory Manual
Course: 410
Subject: Practical-
Course: 410
Subject: Practical-
Molecular Biology & Biotechnology

Prepared By
Md. Imran Nur Manik
M.Pharm
Departmnt of Pharmacy
Uniersity of Rajshahi
Md. Imran Nur Manik
M.Pharm
Departmnt of Pharmacy
Uniersity of Rajshahi
Rajshahi-6205, Bangladesh
Available at
Essential Pharma documents
www.pharmacydocs.blogspot.com
Essential Pharma documents
www.pharmacydocs.blogspot.com
INDEX
Serial No.
|
Date
|
Name of the experiment
|
Page No.
|
01
|
22.02.12
|
Isolation of plasmid DNA from E. coli and determination of concentration by UV-spectrophotometer.
|
2 – 6
|
02
|
25.02.12
|
Estimation of protein concentration from supplied sample by Lowry method.
|
7 – 9
|
Experiment No. 01
|
Date: 22.02.12
|
Name of the experiment: Isolation of plasmid DNA from E. coli and determination of concentration by UV-spectrophotometer.
| |
Principle:
A plasmid is a circular DNA molecule external to the bacterial chromosome, that is able to replicate in its own and distribute its daughter molecules to daughter cells.
Plasmid can be isolated by a varying of methods, many of which rely on the differential denaturation and reannealing of plasmid DNA compared to chromosomal DNA. One commonly used technique in molecular biology is alkaline lysis. This method essentially relies on bacterial lysis by sodium hydroxide (NaOH) and sodium dodecyl sulphate (SDS), followed by neutralization with a high concentration of low-pH potassium acetate. This gives selective precipitation of the bacterial chromosomal DNA and other high molecular weight cellular components. The plasmid DNA remains in suspension and is precipitated with isopropanol.
As nucleic acid shows absorbance at UV-range, the concentration determination is done through the spectrophotometer at 260 nm and it should be taken into consideration that:
1 OD260 = 50 μg/mL
The equation used for calculating the concentration is as follows:
Concentration of nucleic acid = Absorbance × Dilution factor ×50
Reagents:
- For preparation of Luria broth:
- Yeast extract
- NaCl
- Tryptophan
- Distilled water
- For culturing the bacterial cells:
- Luria broth b. E. coli colony
- For isolation of plasmid DNA:
- Solution 1 e. 5 M NaCl
- Solution 2 f. 95% ethanol
- Solution 3 g. Distilled water
- Tris-EDTA (TE) h. Overnight culture of E. coli
Preparation of reagents:
Reagent
|
Ingredient
|
Content
|
Solution 1:
|
50 mM glucose:
25 mM Tris-HCl:
10 mM EDTA:
Water:
|
Per 500 ml
9 ml 50% glucose
12.5 ml 1 M Tris-HCl
10 ml 0.5 M EDTA
Up to 500 ml
|
Solution 2:
|
1% SDS:
0.2 N NaOH:
Water:
|
Per 500 ml
50 ml 10% SDS
100 ml 1N NaOH
Up to 500 ml
|
Solution 3:
|
3 M K+:
5 M Acetate:
Water:
|
Per 500 ml
300 ml 5 M potassium acetate
57.5 ml glacial acetic acid
Up to 500 ml
|
Tris-EDTA (TE):
|
10 mM Tris-HCl:
1 mM EDTA:
Water:
|
Per 100 ml
1 ml 1 M Tris-HCl
0.5 ml 0.5 M EDTA
Up to 100 ml
|
Apparatus:
- For preparation of Luria broth:
- Bottle b. Autoclave
- For culturing the bacterial cells: The E. coli is grown for overnight at 37°C at Luria broth media. Following apparatus are required for this purpose:
- Sterile toothpick
- Sterile tweezers
- Alu-foil
- Autoclave
- Pressure cooker
- Conical flasks
- For isolation of plasmid DNA:
- 1.5 ml microfuge tube
- Centrifuge test tube
- Micropipette
- Vortex machine (Biocraft, Biocraft & Scientific industries)
- Syringe with 26 needle
- UV-spectrophotometer (Shimadzu, Model: UV-1200)
- Centrifuge machine (800 centrifuge machine)
Procedure:
- Preparation of Luria broth: For 1000 ml volume of Luria broth; 5 gm of yeast extract, 10 gm of NaCl, 10 gm of tryptophan and 1000 ml distilled water were taken in a bottle. Then the prepared solution was autoclaved for 15 minutes.
- Preparation of E. coli solution: In this method; we used a sterile toothpick, a sterile tweezer and a culture of E. coli. The toothpick was sterilized by wrapping it in Alu-foil and placing in an autoclave for 20 minutes at a pressure of 15 Ibs and a temperature of 121°C for. A match was used to flame the end of a pair of tweezers. The sterile toothpick was picked up with sterile tweezer, a colony or section of E. coli was put on the toothpick and dropped into the bottle of Luria broth. The bottle was capped loosely and incubated at 37°C.
- Isolation of plasmid DNA from E. coli:
- 1.5ml of bacterial culture was transferred to a microcentrifuge tube. The tube was then centrifuged at 4,000 rpm for 5 minutes and consequently the bacterial cells were pilleted. The supernatant of the tube was discarded carefully.
- Process of step-1 was repeated for 3 times in the same tube. The purpose of this step is to increase the starting volume of cells so that more plasmid DNA can be isolated.
- 250 μl of solution-1 was added to cell pellet and resuspended cells as much as possible by using disposable syringe.
- About 200/250 μl solution-2 was added. The solution was mixed gently by inverting and rotating the tube for several times. The tube should not be vortexed. After that, the tube was kept at room temperature for 5 minutes, but not more than 5 minutes as it may cause the denaturation of plasmid DNA.
- 350 ml ice-cold solution-3 was then added in the microcentrifuge tube and the solution was mixed gently by inverting tube for 6-8 times.
- The tube was then centrifuged for 10 minutes. Two layers were formed, uppers layer is plasmid DNA and lower layer is the mixture of chromosomal DNA, protein, cell debris, SDS etc. The supernatant was transferred to a fresh microcentrifuge tube by using clean disposable transfer pipet. Care was taken as white precipitate might not take during the transfer.
This fractionation step separates the plasmid DNA from the cellular debris and chromosomal DNA in the pellet.
- The tube was then filled with isopropanol up to the mark. The solution was again mixed gently by inverting and rotating the tube for several times and the tube was then kept at room temperature for 2 minutes.
Isopropanol effectively precipitates nucleic acids, but is much less effective with proteins. A quick precipitation can therefore purify DNA from protein contaminants.
- The tube was then centrifuged for 10 minutes. After centrifugation, a milky white pellet was adhered at the bottom of the tube. The supernatant was decanted without dumping out the pellet.
This fractionation step further purifies the plasmid DNA from contaminants.
- 1 ml of ice-cold 70% ethanol was added. The solution was mixed gently by inverting the tube for several times, then kept at – 20°C for 15 – 30 minutes and after that centrifuged for 10 – 15 minutes. The supernatant was decanted without dumping out the pellet.
Ethanol helps to remove the remaining salts and SDS from the preparation.
- The tube was allowed to dry for ~5 minutes and after that, 50 μl TE was added to the tube. This was the isolated plasmid DNA solution.
- The DNA concentration was determined by measuring the absorbance at 260 nm (1 OD260 = 50 μLg/ml). For this μL sample solution was taken into absorbance cell which was diluted with μL of distilled water.
Calculation:
The absorbance data were as follows:
Blank solution =
Sample solution: For nucleic acid, At λ260 =
For proteins, At λ282 =
Ratio = 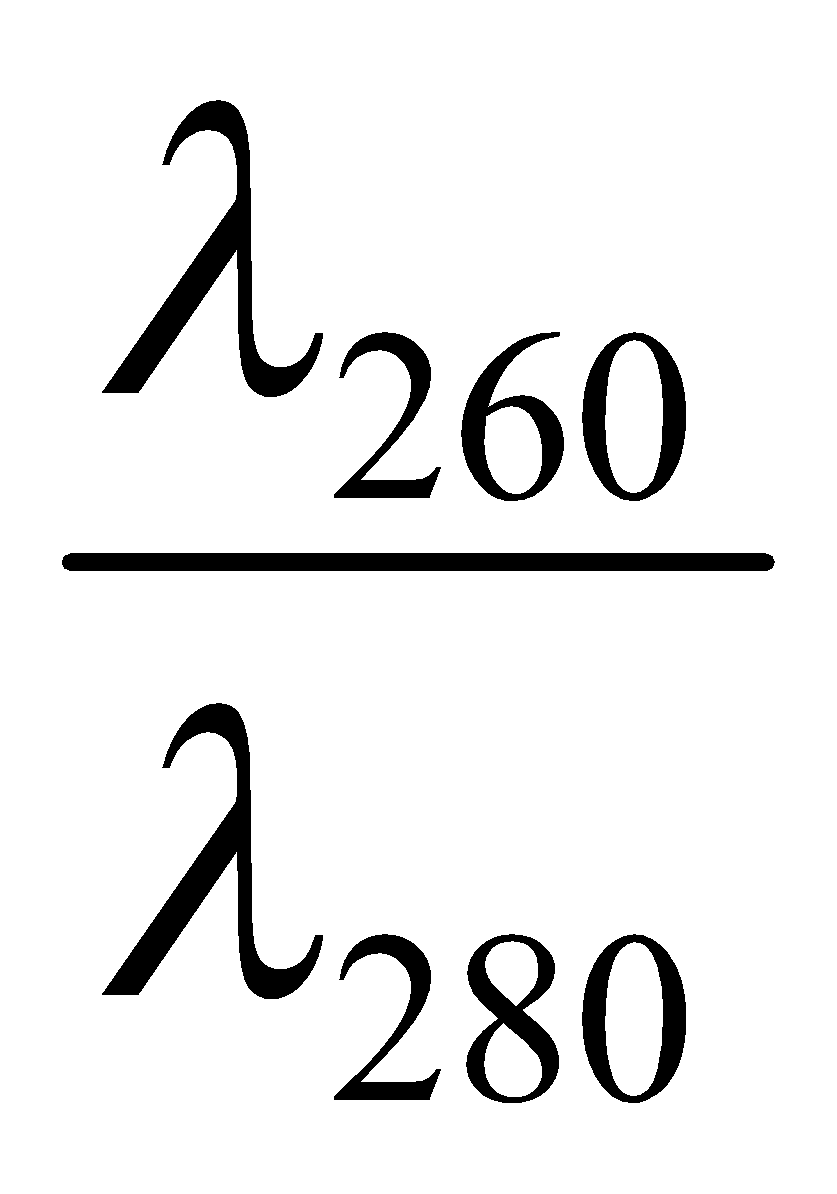 =
=
If the ratio is equal to 1.8 the DNA is pure, otherwise protein complexed DNA (i.e. impure).
Dilution factor:
Dilution factor is the ratio between total amount of solution and amount of sample solution.
That is, Dilution factor = 
In this experiment, μl sample solution was mixed with μL distilled water. So, the total amount of solution was μl.
Thus, Dilution factor = 
We know that, 1 OD260 = 50 μg/ml
So, the concentration of nucleic acid = Absorbance × Dilution factor ×50
=
= μg/mL
Result:
The concentration of nucleic acid was μg/mL.
Comments:
Experiment No. 02
|
Date: 25.02.12
|
Name of the experiment: Estimation of protein concentration from supplied sample by Lowry method.
| |
Principle:
The Lowry method was developed by Lowry in 1957. This is relatively more effective method. The Lowry method is based on taking the absorbance of supplied sample, which is diluted with distilled water and treated with alkaline solution (pH 10) such as reagent A, B and C etc. solution and followed by Folin phenol reagent (Sodium tungstate-molybdate and phosphate or Phosphomolybdotungstate) solution at 750 nm and then determining the concentration of protein from standard calibration curve.
Protein reacts with Cu2+ ion of the alkaline solution to form a colour complex (bluish yellow) which then reacts with constituents of folin phenol solution to form another coloured complex. So, the absorbance is taken at visible range.
Another alternative method is Branford’s method.
Reactions:
Step-1: At first, protein reacts with Cu2+ in alkaline condition at pH 10 and form protein-Cu2+ complex which is somewhat bluish yellow in colour.
Protein + Cu2+ ⎯⎯→ Protein – Cu2+ complex
Step-2: In this step, folin phenol reagent reacts with protein – Cu2+ complex, reduces to polymolybdenum and forms a complex of blue colour. This blue colour depends on the ratio of tryptophan, tyrosine and cysteine concentration in the ptrotein.
Folin phenol reagent + Protein-Cu2+ complex ⎯→ Reduced product of
polymolybdenum (blue)
The intensity of colour change is directly proportional to protein concentration. Finally absorbance of sample solution is taken at a 750 nm and by measuring the absorbance, the concentration of protein is determined.
Apparatus:
- 100 ml beaker iv. Pipette
- 100 ml volumetric flask v. 100 ml measuring cylinder
- Test tubes vi. Spectrophotometer
Reagents:
- Reagent A = 2% Na2CO3 in 0.1 N NaOH
- Reagent B = 1% CuSO4 . 5H2O
- Reagent C = 1% Na-K tartarate
- Reagent D = A : B : C = 100 : 1 : 1
- Reagent E = 2 N Folin phenol reagent : H2O = 1 : 1
- Preparation of reagent A: It is a solution of 2% Na2CO3 in 0.1 N NaOH solution. It was prepared by dissolving 4 gm Na2CO3 and 0.8 g NaOH in some portion of distilled water in a 200 ml volumetric flask. Then the volume was adjusted up to 200 ml by adding distilled water.
- Preparation of reagent B: It was prepared by dissolving 0.5 gm CuSO4 . 5H2O in 50 ml distilled water.
- Preparation of reagent C: It was prepared by dissolving 1 gm Na-K tartarate in some portion of distilled water and after dissolving the volume was adjusted up to 100 ml by adding distilled water.
- Preparation of reagent D: It was prepared by mixing 100 ml reagent A, 1 ml reagent B and 1 ml reagent C.
- Preparation of reagent E: 2N folin phenol reagent is commercially available. Reagent-E was prepared by taking 5 ml folin phenol solution and mixing with 5 ml distilled water in 1:1 ratio.
- Preparation of stock solution: A standard Bovine serum albumin (BSA) was prepared by dissolving 10 mg BSA in 50 ml distilled water. This solution was used as standard stock solution. The concentration of this solution was 200 μg/ml.
- Preparation of sample solution: From 2 ml supplied sample, 1 ml was taken in a test tube and 5 ml of reagent-D was added and vortexed for 1 minute. Then 0.5 ml of reagent-E was added and again vortexed for 1 minute. This solution is sample solution.
viii. Preparation of blank solution: 1 ml distilled water was taken in a test tube and 5 ml of reagent-D was added and vortexed for 1 minute. Then 0.5 ml of reagent-E was added and again vortexed for 1 minute. This solution is blank solution.
Procedures:
- 0.2 ml, 0.4 ml, 0.8 ml, 1.2 ml and 1.6 ml stock solution were taken in five different test tubes and numbered as standard-1, 2, 3, 4 and 5. The volume of each test tube was adjusted to 2 ml by adding 1.8 ml, 1.6 ml, 1.2 ml, 0.8 ml and 0.4 ml distilled water respectively. Thus, the concentration of solutions was 20 μg/ml, 40 μg/ml, 80 μg/ml, 120 μg/ml and 160 μg/ml respectively.
- 1 ml solution from each of the 2 ml solution in each test tube was taken in another five different test tubes.
- 5 ml of freshly prepared reagent-D was added in each test tube and each test tube was vortexed for 1 minute.
- 0.5 ml of reagent-E was added rapidly and again each test tube was vortexed for 1 minute.
- All the test tubes were then allowed to stand at room temperature for 30-35 minutes.
- The absorbance of blank, sample and all standard solutions was taken by UV-spectrophotometer at 750 nm.
- Then a standard graph of absorbance versus concentration was plotted for standard solution.
- After that, concentration of test sample was calculated from the standard curve. It was done by plotting absorbance value of sample solution to y-axis of the standard curve and a perpendicular was drawn from the absorbance point of sample to the x-axis.
Calculation:
50 ml stock solution contains = 10 mg protein
1 ,, ,, ,, ,, =  ,, ,,
,, ,,
= 0.2 mg protein
= 200 μg protein
Thus; 0.2 ml stock solution contains = 200 × 0.2 = 40 μg protein
0.4 ,, ,, ,, ,, = 200 × 0.4 = 80 μg protein
0.8 ,, ,, ,, ,, = 200 × 0.8 = 160 μg protein
1.2 ,, ,, ,, ,, = 200 × 1.2 = 240 μg protein
1.6 ,, ,, ,, ,, = 200 × 1.6 = 320 μg protein
Data for protein concentration:
Sample
|
Conc. of protein (μg/ml)
|
Absorbance
|
Conc. of protein in sample
(μg/ml)
|
Blank solution
| |||
Standard-1
| |||
Standard-2
| |||
Standard-3
| |||
Standard-4
| |||
Standard-5
| |||
Sample solution
|
Now from the standard curve of protein concentration, we can easily calculate the protein concentration in supplied sample.
Result:
The concentration of protein in supplied sample was μg/ml.
Comments:
No comments:
Post a Comment